Getting Started
- FishExplorer is a JavaScript-based visualization tool and therefore can be used in any modern browser without installing software. Currently as backend (database server) a server at ZIB is used. In the future it will be possible to connect to other data servers by a login mechanism. At the ZIB database server, currently only the zebrafish reference frame is available. Further data can be made available by sending us the data.
- In this section, you will learn how to
- Download the current project
- Load the project
- Invoke viewers for loading data i.e brain regions, single cells etc
-
When FishExplorer is started, a window like the one shown below appears on the screen, The top region
page is divided into three main parts.
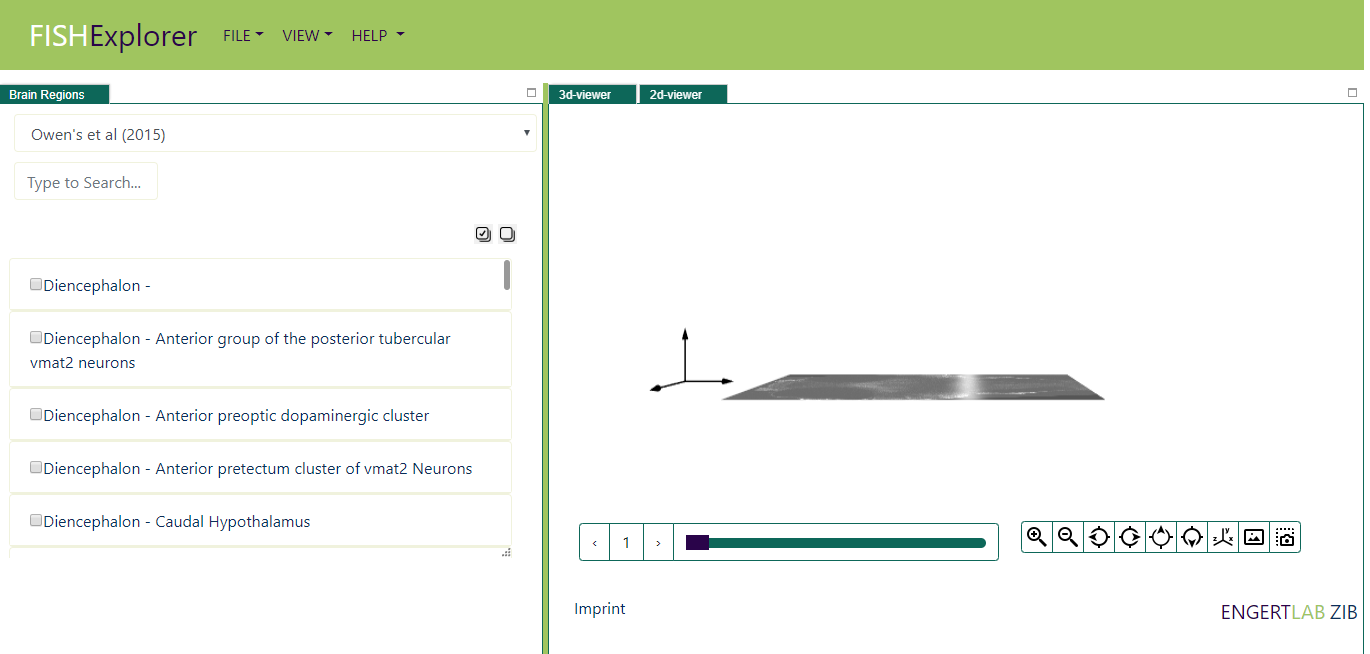
-
File
It consist of two parts:
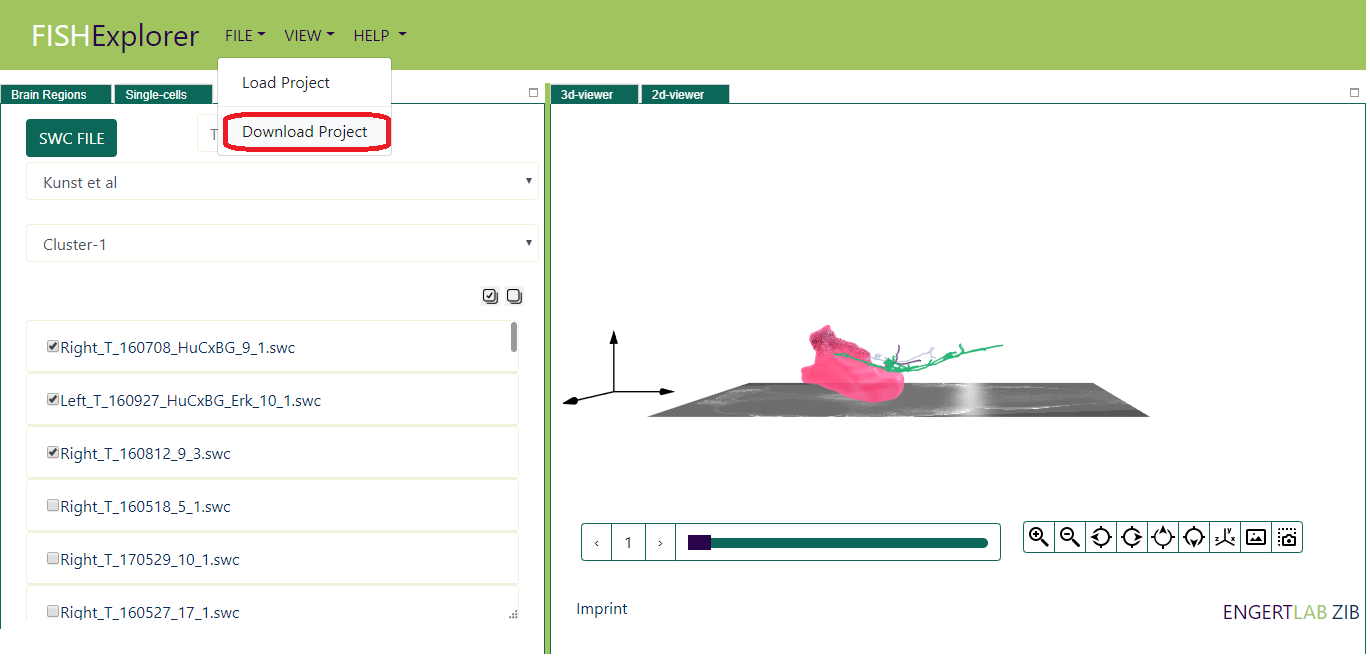
- Download Project: allows users to store current state of the project locally usings metadata about brain regions, cells and morphologies
(as show in picture below)
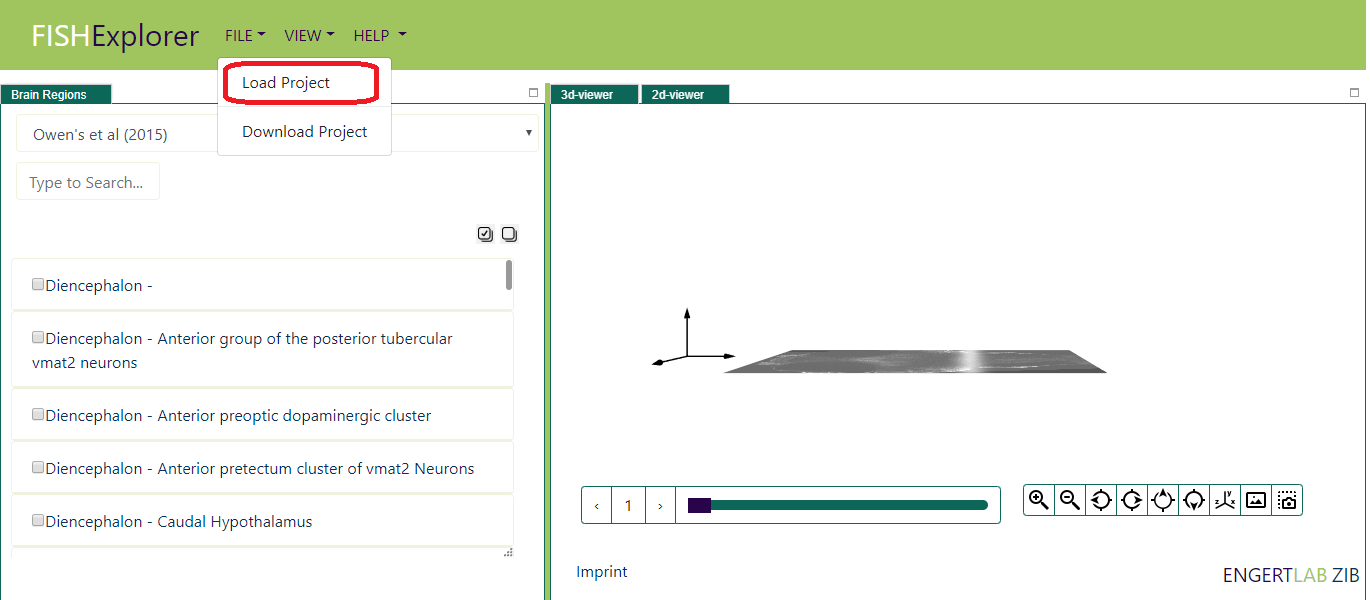
- Load Project allows users to load locally stored project in the platform. As a testcase, user can load
this project file (as show in picture below)
The list of brain regions, cells and morphologies mentioned in project file will be loaded (as show in picture below)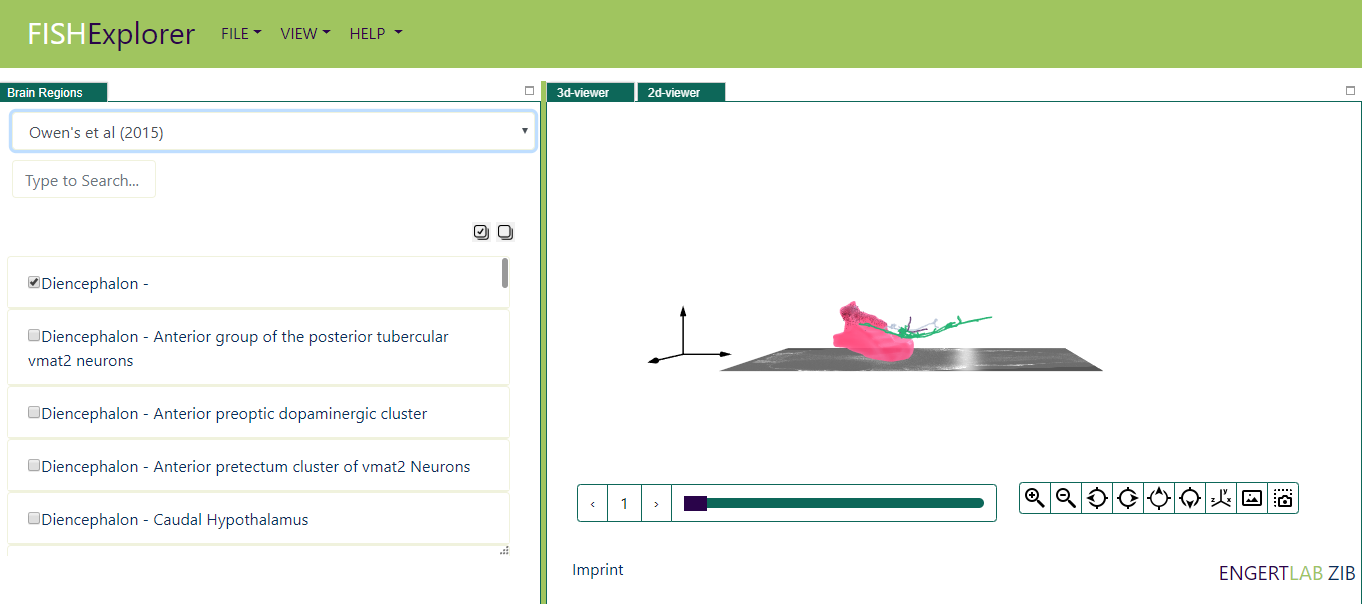
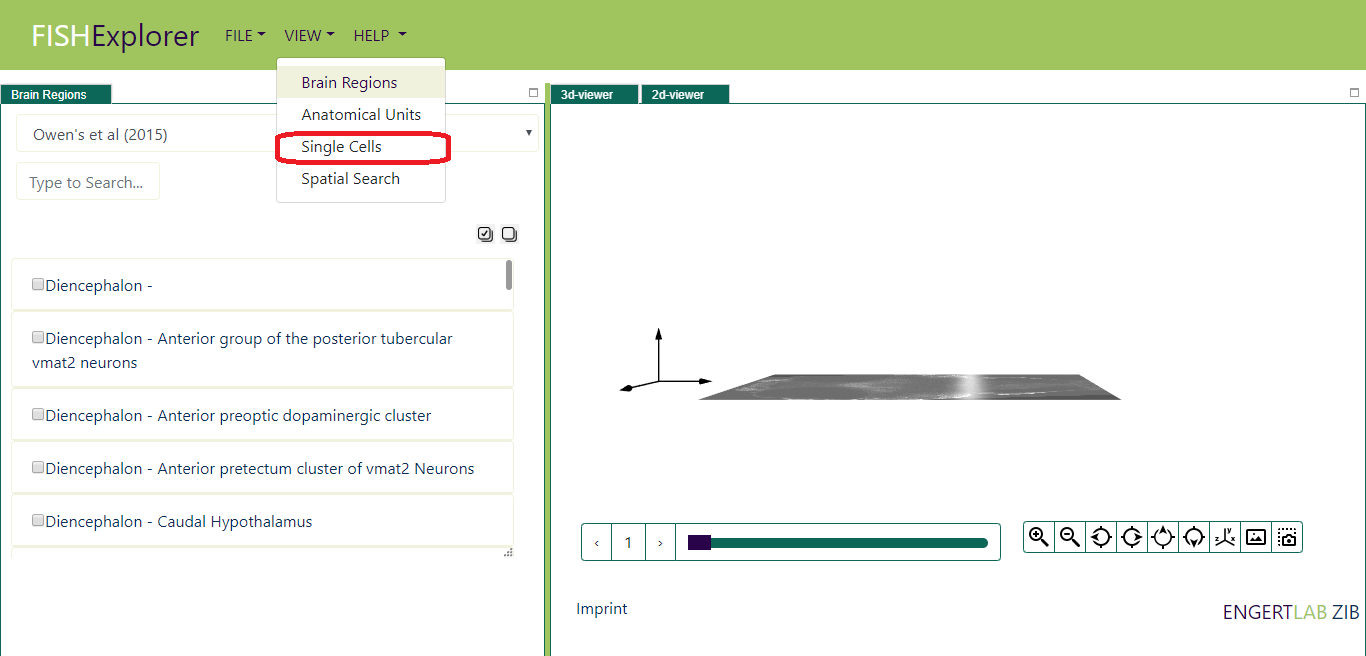
Click the view button, a list of viewers like the one shown below appears on the screen.
- Download Project: allows users to store current state of the project locally usings metadata about brain regions, cells and morphologies
(as show in picture below)
- Click the link "Single Cells" (encircled red in the image above), Single cell viewer will appear on screen
(as shown in the image below).
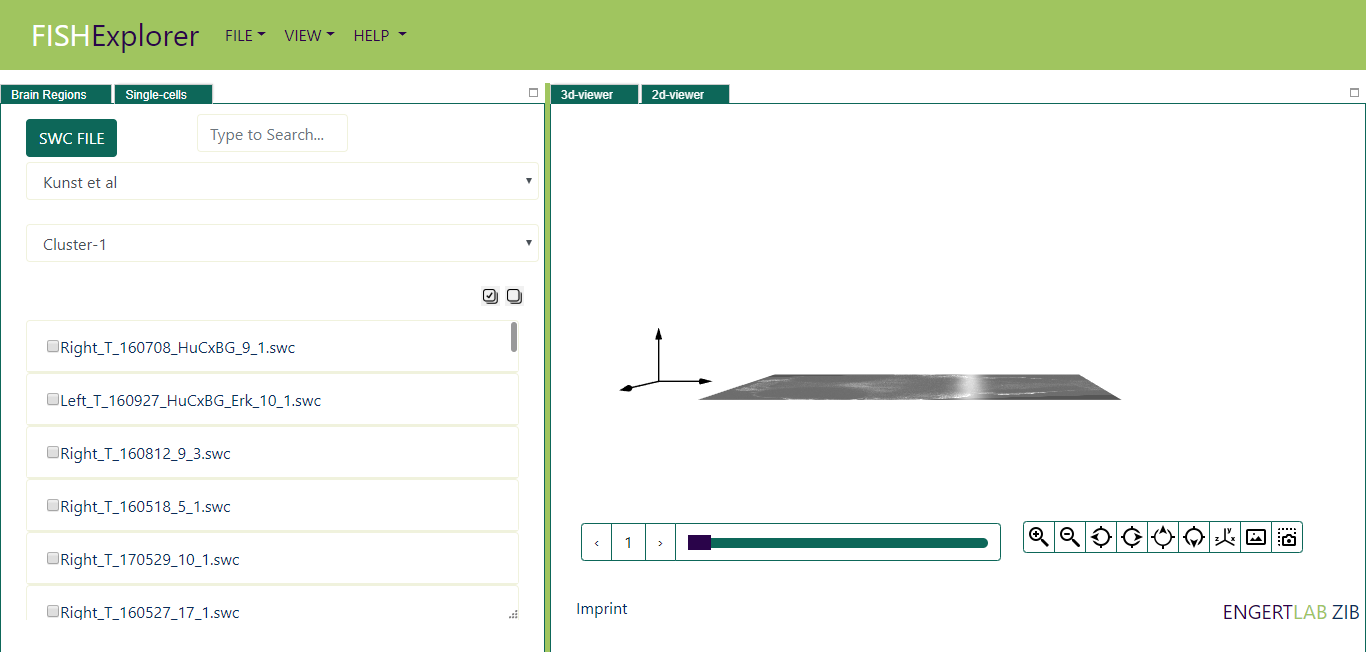
The default selected dataset is Kuns et al, list of predefined clusters for the selected dataset will appears
on the screen (encircled red in the image below)
 Help
Help
It contains some links to help of FishExplorer.